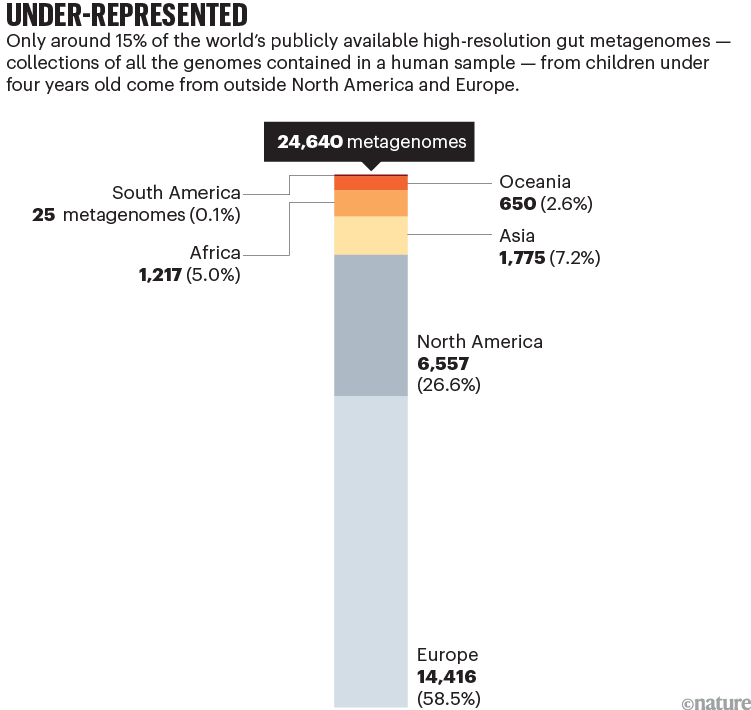
Less than 15% of the global population lives in Europe or North America. Yet more than 70% of published human microbiome data — on the collections of bacteria, fungi and viruses that live on and in our bodies — comes from European and North American populations1. Around 85% of the 25,000 high-resolution gut metagenomes from children under four that are publicly available come from individuals living in these wealthy regions (see ‘Under-represented’). In this context, metagenomes are collections of all the genomes contained in a faecal, skin or other human sample.
Likewise, investigators are beginning to explore the microbiota as a therapeutic target for various diseases that are common in high-income countries, such as metabolic disorders, cancer and inflammatory bowel disease. Much less attention is being given to how the microbiota affects conditions such as malnutrition and infectious diseases that disproportionately affect people living in low- and middle-income countries (LMICs).
This must change. It is now clear that the gut microbiota — the most studied of the human microbial communities — of children and adults can differ markedly depending on where people live. So the development of safe and effective microbiome-based therapeutics for those living in the world’s poorer regions depends on microbiome data being collected from these areas.
Sources: H. P. Browne et al.; data from NCBI/MGnify
To explore how microbiome research could be accelerated globally, last year, four of us (H.P.B., N.T.I, M.O. and C.T.) helped to organize a two-day workshop at the Wellcome Genome Campus in Cambridge, UK. Funded by Wellcome Connecting Science and the Bill & Melinda Gates Foundation, this event — which all eight of us attended — brought together leaders in early-life microbiome research as well as early-career researchers from 23 countries.
All 39 of the researchers, funders and industry representatives attending the event agreed that microbiome science has enormous potential to improve people’s health globally — especially the health of children. We also agreed that achieving this goal will require the collection of data from many diverse populations worldwide and the training of researchers in LMICs. It will also need the development of local infrastructure to analyse specimens and conduct clinical studies, and long-term collaborations involving researchers from LMICs, and from institutions and corporations in Europe and North America — which currently lead microbiome research.
Good for health
We are born without a microbiota, but our bodies are rapidly colonized by diverse microbes — from our mothers, the local environment and other people in our social networks2,3. Sequencing studies indicate that in healthy individuals, the human gut microbiota develops in a definable way, whereby the arrival of one species influences which species colonizes the gut next and so on.
This ‘ecological succession’ begins at birth with the transfer of bacteria from the mother to the infant during labour and then through skin-to-skin contact and br4eastfeeding. Some species of Bifidobacterium, for example, are highly adapted for colonizing the guts of infants because they use carbohydrates found in mucus and breast milk. When an infant stops breastfeeding, some of these Bifidobacterium species essentially disappear, enabling other bacteria, such as those in the spore-forming Lachnospiraceae family, to establish themselves.
In breaking down the carbohydrates in the gut, Bifidobacterium species produce various metabolites that are beneficial to infants, such as organic acids, B vitamins, neurotransmitters and proteins that modify the immune system4,5. Thus, the healthy co-development of humans and their gut microbiota is key to healthy growth. In fact, there is growing evidence that disrupting the assembly of the gut microbial community after birth — whether through pathogen infection or undernutrition — is linked to wasting (in which a child’s weight is low for their height) and stunting (in which a child’s height is low for their age), as well as other physiological conditions6.
Peolple at a clinic in Cité Soleil, Port-au-Prince, receive treatment for cholera.Credit: Richard Pierrin/AFP/Getty
The microbiome-based therapeutics that are currently being developed and tested in European and North American countries include ‘next generation’ probiotic formulations. Here, investigators characterize what is typical for an individual and then work out whether an aberration in that person’s microbiota is the cause or effect of a given disease. They then identify therapeutic targets and develop microbiome-directed therapeutic candidates for testing in humans.
But to complicate things, the mix of microbes in the gut and the functions of individual species and subspecies change throughout a person’s life7. What’s more, which particular bacterial species (or subspecies) are present at any one time varies depending on where an individual lives8. The Bifidobacterium longum subspecies infantis (B. infantis) that is commonly found in young children in Africa, for instance, is largely absent from the guts of children living in the United States9.
Location matters
When it comes to LMICs, evidence is now emerging that the gut microbiome could offer leads for therapeutics for some of these nations’ biggest public-health threats.
Take undernutrition, which is associated with nearly half of all deaths of children under five — 3.1 million annually10. In studies using gnotobiotic mice (which have a defined microbiota and are reared in sterile conditions), researchers have in recent years identified various food ingredients that promoted the growth of certain bacterial strains that were under-represented in children with malnutrition11,12. In subsequent clinical trials, giving a gut-microbiota-directed food formulation containing these ingredients to malnourished children aged 12–18 months in Bangladesh increased the growth of the children. It also altered their microbiomes so that they more closely resembled those found in healthy Bangladeshi children13. A common ready-to-use supplementary food given to children in the same clinical trial did not achieve the same health effects, despite containing more calories than the microbiota-guided food ingredients.
How our microbiome is shaped by family, friends and even neighbours
Because of the genetic and phenotypic diversity of people’s microbiomes, findings made in Europe and North America will not necessarily apply to other regions — and treatments developed in wealthy countries might not help people in poorer countries who could benefit from them the most.
In a 2022 study, for instance, researchers gave Bangladeshi infants aged 2–6 months with severe acute malnutrition a probiotic strain that had been cultured from a donor in the United States14. The infants in the study had lower levels of B. infantis in their intestines than did healthy infants from Bangladesh.
The probiotic improved the growth rates of the malnourished infants and reduced inflammation in their guts. But the levels of colonization achieved in the study were 10- to 100-fold lower than those found in healthy infants14. On the basis of follow-up studies in gnotobiotic mice, the researchers postulated that this reflected the fact that the introduced US-derived B. infantis strain lacked certain genes that are present in strains cultured from Bangladeshi infants. Infants in Bangladesh commonly consume certain plants, and strains cultured from their guts contain genes that are involved in the metabolism of the plant carbohydrates.
Such variation between countries is likely to be important in other contexts, too.
As well as helping people to obtain certain nutrients, the intestinal microbiota protects them against infection by competing with pathogens for nutrients or by producing antimicrobial molecules that destroy them. Various studies have shown that in different settings, beneficial gut microbes encounter different species and strains of pathogens15. So the gut microbes found in communities living in one place might not have evolved effective ways to compete with pathogens that are endemic elsewhere.
Likewise, the resistance of gut pathogens to antibiotics varies with geography, depending in part on local health procedures and regulations around the use of antibiotics. And vaccines developed to protect people against pathogens in one area might not work so well in other areas. Take rotaviruses, one of the most common causes of diarrhoeal disease among infants and young children. The rotavirus vaccine is up to 90% effective at preventing disease in high-income countries, but only up to 50–60% effective in African and South Asian countries. This could be because of different immune responses to the vaccine owing to different microbiota compositions in children16.
In short, researchers trying to develop therapeutics to tackle malnutrition or infectious diseases, will need to conduct studies wherever the intervention will be used.
A worldwide push
So, how can microbiome research be accelerated in LMICs?
Currently, numerous barriers are hampering progress. Researchers in LMICs often lack the transport services, freezers and reliable power supplies needed to obtain samples and rapidly freeze them at extremely low temperatures (−80 °C or lower). Freezing samples ensures the integrity of DNA and RNA for sequencing, and bacterial viability for culturing. LMICs often lack the computational resources needed to analyse and store vast data sets. They also don’t have access to sealed anaerobic cabinets that are required for culturing oxygen-intolerant gut bacteria, archiving the cultured organisms, sequencing their genomes and characterizing their growth requirements and metabolic outputs. These countries also often lack the facilities needed to perform studies in gnotobiotic mice.

Microbiome research could help to address some major public-health threats in LMICs.Credit: Matthew Midgley/Wellcome Sanger Institute
Added to this are the relatively high costs of reagents and the long waiting times to receive them from manufacturers owing to challenges around customs and supply chains. Perhaps most importantly, in many LMICs, people lack the training and expertise required to develop a multidisciplinary workforce that can conduct microbiome studies.
Three steps, however, could lift many of these barriers.
Establish regional centres of excellence. Currently, microbiome projects tend to take samples from donors for, at most, a year or two. The establishment of regional centres of excellence dedicated to microbiome research would enable researchers in LMICs to conduct long-term sampling of the microbial ecology in a population (over decades) — and to map and preserve strain-level microbial diversity. Such centres — which could be funded by local governments as well as by organizations such as Wellcome and the Bill & Melinda Gates Foundation — could drive the training of researchers from other regions. These centres could also provide a hub for sharing data, expertise, policies and procedures — as the African Centre of Excellence for Genomics of Infectious Diseases based at Redeemer’s University in Ede, Nigeria, does for infectious diseases.
The Cape Town Statement on fairness, equity and diversity in research
Initially, these centres could be established in universities or in research institutes that already have some infrastructure in place, such as the Kenya Medical Research Institute in Nairobi, the International Centre for Diarrhoeal Disease Research in Dhaka, or the Aga Khan University in Karachi, Pakistan. But many other factors should inform decisions about where to establish such centres — including the impacts of malnutrition or infectious diseases, such as cholera or typhoid fever, on a particular area, and the local governmental policies that regulate sampling.
Develop microbial culture collections. After obtaining informed consent from caregivers, including for long-term use of the samples, researchers in LMICs need to establish curated microbial culture collections — particularly from children.
Several examples provide guidance on how researchers can engage stakeholders, including study participants. In 2015, for example, researchers from the United States and Peru, who wanted to characterize the gut microbiomes of Indigenous people in Peru, worked closely with local communities. During this time, they reviewed study procedures with community leaders, held public meetings for consultation, provided opportunities for community members to observe the scientists processing samples, and explained initial results17.
By culturing individual strains from a sample, and then identifying what genes each isolate carries, researchers could develop microbial reference genomes specific to the local population. By comparing these genomes with those generated from other populations, scientists could then identify population-specific microbial adaptations, such as those linked to diet or breastfeeding patterns. And by linking genomes back to cultured isolates and conducting experiments on them, the researchers could unpick the mechanisms underlying a particular adaptation. High-quality reference genomes could even be used to improve the taxonomic resolution of metagenome profiling, in which researchers sequence millions of short stretches of DNA from a gut sample to assess what microbial species and subspecies are present18.
These curated culture collections linked to high-quality reference genomes and metagenomes would provide a valuable resource for microbiome research, and ultimately pave the way for the development of microbiome-based therapeutics.
Foster collaboration. Robust networks between the better-resourced academic laboratories in Europe and North America and researchers in LMICs will be crucial. Such collaborations should span decades rather than a few years — particularly because recruiting participants for multi-year studies, collecting and analysing the microbiome data and associated metadata produced, and then validating the findings using animal models and clinical trials can take more than ten years.
The workshop at the Wellcome Genome Campus connected researchers from various countries with each other as well as with funders. But besides events of this kind, exchange programmes and other initiatives could help scientists from LMICs get the training they need to become leaders in microbiome research in their own countries. Targeting more research grants to specific countries would help support trainees to continue their research in their home countries.
Efforts to ensure that genomic data and analysis tools are shared and used fairly among all collaborators have increased in the past few years. We welcome this. Yet, international legislation such as the 2010 Nagoya Protocol, which strives to ensure equitable access to genetic resources, can — in our experience — make it harder for scientists working in LMICs to engage in international collaborations19.
Some countries that have adopted the Nagoya Protocol fail to provide researchers with guidance on what steps to take in practice. Also, working with lawyers to draw up agreements for the transfer of materials from one country to another can take months and cost thousands of dollars. Going forward, more transparent, standardized and efficient, but ethically grounded, procedures are required.
Benefits for all
So long as microbiome-based therapeutics are designed, manufactured and delivered by organizations outside LMICs, many of the economic and societal benefits resulting from these treatments probably won’t reach the communities that need them most. Conversely, a better understanding of the composition, functions and evolutionary forces at play in people’s gut and other microbiotas — from diverse settings — could improve long-term health outcomes for everyone.
Source link